Introduction.
Multiple myeloma (MM) is an incurable plasma cell malignancy with a growing list of anti-MM therapeutics. However, the development of predictive biomarkers has yet to be achieved for nearly all MM therapeutics. Selinexor (SELI), a nuclear export inhibitor targeting exportin 1 (XPO1), has been approved with dexamethasone (DEX) in penta-refractory MM. Clinical studies investigating promising SELI- 3 drug combinations are ongoing. Here, we have investigated potential synergistic combinations of SELI and anti-MM agents in terms of ex vivo sensitivity, as well as paired RNAseq and WES to identify companion biomarkers.
Methods.
MM cells isolated from fresh bone marrow aspirates were tested for drug sensitivity in an organotypic ex vivo drug sensitivity assay, consisting of co-culture with stroma, collagen matrix and patient-derived serum. Single agents were tested at 5 concentrations, while two-drug combinations were tested at fixed ratio of concentrations. LD50 and area under the curve (AUC) were assessed during 96h-exposure as metrics for drug resistance. Drug synergy was calculated as a modified BLISS model. Matching aliquots of MM cells had RNAseq and WES performed through ORIEN/AVATAR project. Geneset enrichment analysis (GSEA) was conducted using both AUC and LD50 as phenotypes for single agents and combinations. Both curated pathways (KEGG and cancer hallmarks) and unsupervised gene clustering were used as genesets. Student t-tests with multiple test correction were used to identify non-synonymous mutations in protein coding genes associated with single agent or combination AUC.
Results.
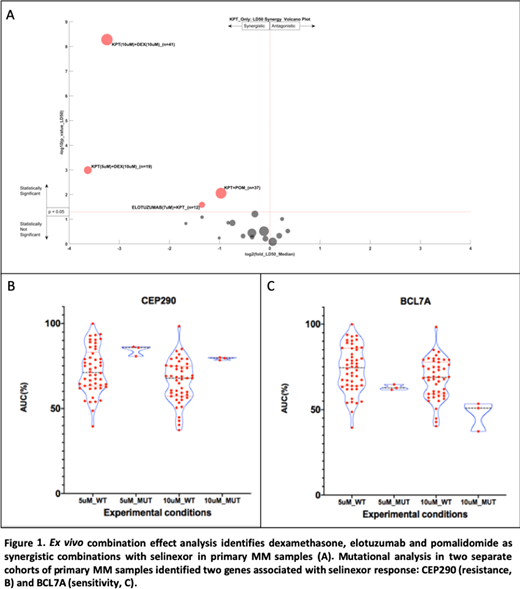
For this analysis, a cohort of specimens from 103 patients (48% female, 4% Hispanic, 11% African American) was tested with SELI and/or DEX. with a median of 2 lines of therapy (0-12). A smaller cohort of 37 have been examined with SELI, pomalidomide (POM), elotuzumab (ELO) and daratumumab (DARA). Within this cohort we observed synergy between SELI and DEX, POM and ELO as shown in Figure 1. The volcano plot illustrates the number of samples, maximum drug concentration, as well as magnitude (x- axis) and significance (y- axis) of synergy. Although the SELI-DARA combination trended toward synergy, statistical significance was not achieved.
To identify molecular mechanisms and biomarkers associated with sensitivity to SELI and SELI- combinations, we investigated paired RNAseq and WES with ex vivo sensitivity. Initially, we conducted GSEA on two cohorts of primary MM samples tested with SELI alone at 5µM (n=53) and 10µM (n=50). Cell adhesion (KEGG CAMS), inflammatory cytokines (KEGG ASTHMA), and epithelial mesenchymal transition (HALLMARK EMT) were associated with resistance in both cohorts, while the HALLMARK MYC TARGETS was associated with sensitivity (FWER p<0.05). Mutational analysis identified 46 gene mutations associated with SELI resistance and 100 associated with sensitivity at 5µM, and 87 and 27 mutations associated with SELI resistance and sensitivity, respectively, at 10µM. Two gene mutations were identified in both cohorts: BCL7A, involved in chromatin remodeling, was associated with sensitivity and CEP290, a microtubule binding protein, associated with resistance (p<0.05). Analysis of both gene sequences (NetNES 1.1) identified nuclear export signal (NES) residues suggesting these may be XPO1 cargo. Additionally, translocation t(11;14) was associated with SELI resistance in the 5µM cohort (p=0.037). The completed set of 50 specimens ex vivo, RNAseq and WES analysis will be mature and updated for the potential presentation at ASH.
Conclusions.
We observed ex vivo synergy between SELI and DEX, POM and ELO. Molecular analysis of matched ex vivo drug sensitivity, transcriptome and mutational profile identified environment-mediated drug resistance pathways positively correlated with SELI single agent resistance, as well as MYC regulated genes associated with ex vivo sensitivity. We also identified a list of mutations associated with SELI drug resistance and sensitivity, with special emphasis on two novel NES-containing genes, CEP290 and BCL7A. The next step of this project is to analyze transcriptional and mutational patterns associated with ex vivo synergy in the combinations here described, as putative biomarkers for future clinical investigation.
Shain:Amgen: Speakers Bureau; Adaptive: Consultancy, Honoraria; Karyopharm: Research Funding, Speakers Bureau; Celgene: Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; GlaxoSmithKline: Speakers Bureau; Janssen: Honoraria, Speakers Bureau; BMS: Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Sanofi/Genzyme: Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Takeda: Honoraria, Speakers Bureau; AbbVie: Research Funding. Kulkarni:M2GEN: Current Employment. Zhang:M2GEN: Current Employment. Hampton:M2GEN: Current Employment. Argueta:Karyopharm: Current Employment. Landesman:Karyopharm Therapeutics Inc: Current Employment, Current equity holder in publicly-traded company. Siqueira Silva:AbbVie: Research Funding; NIH/NCI: Research Funding; Karyopharm: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.